Publications
Advances in recent years have brought an abundance of data regarding the molecular level details of biology, yet there is still often an absence of understanding in how molecular level phenomena contribute to health and disease. Our work strives to connect molecular structure and function with macroscale observations to develop computational tools for thereapeutic design.
Highlighted

Reassessing enzyme kinetics: Considering protease-as-substrate interactions in proteolytic networks
Proceedings of the National Academy of Sciences
·
24 Jan 2020
·
doi:10.1073/pnas.1912207117
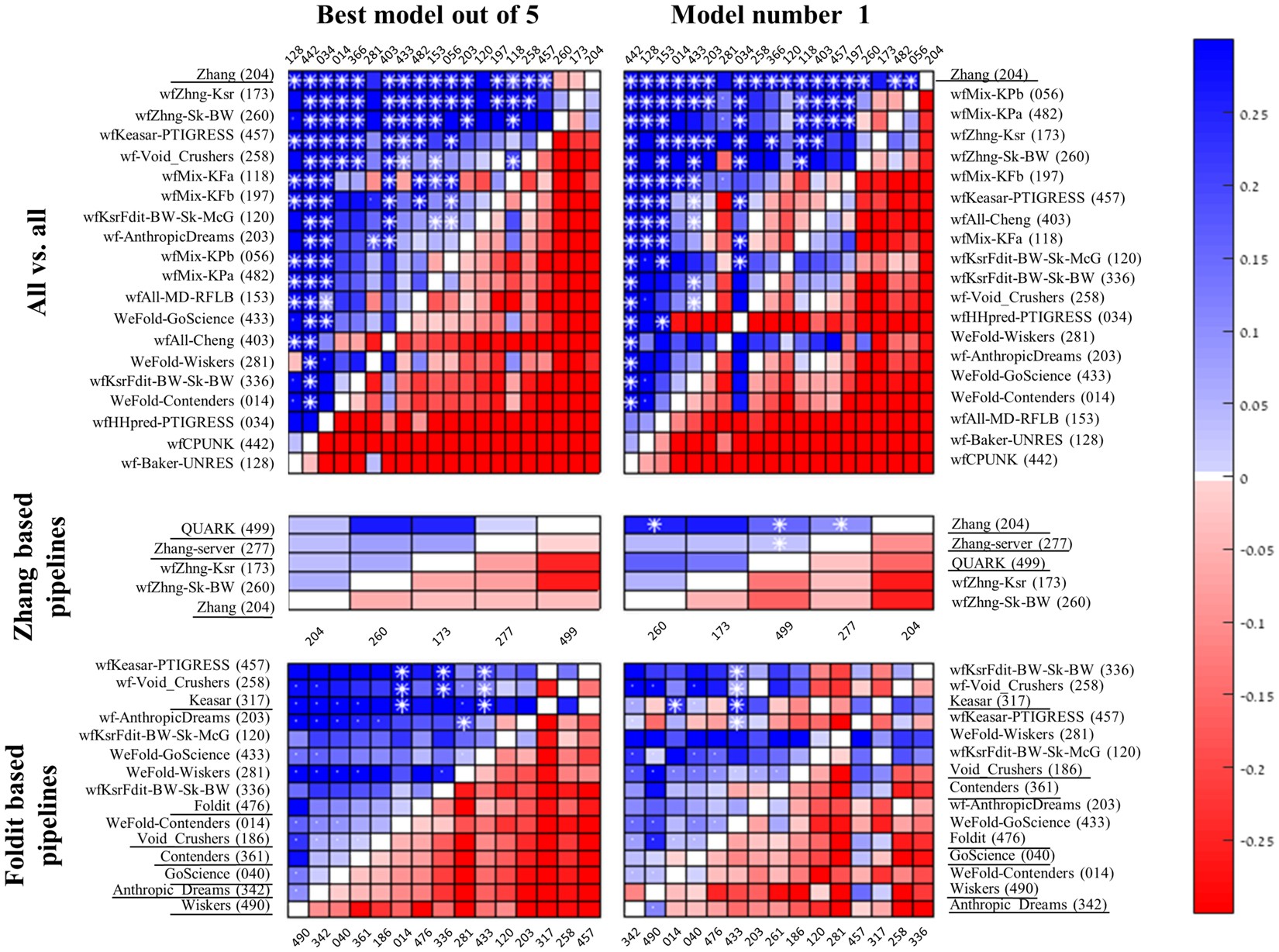
An analysis and evaluation of the WeFold collaborative for protein structure prediction and its pipelines in CASP11 and CASP12
Scientific Reports
·
02 Jul 2018
·
doi:10.1038/s41598-018-26812-8
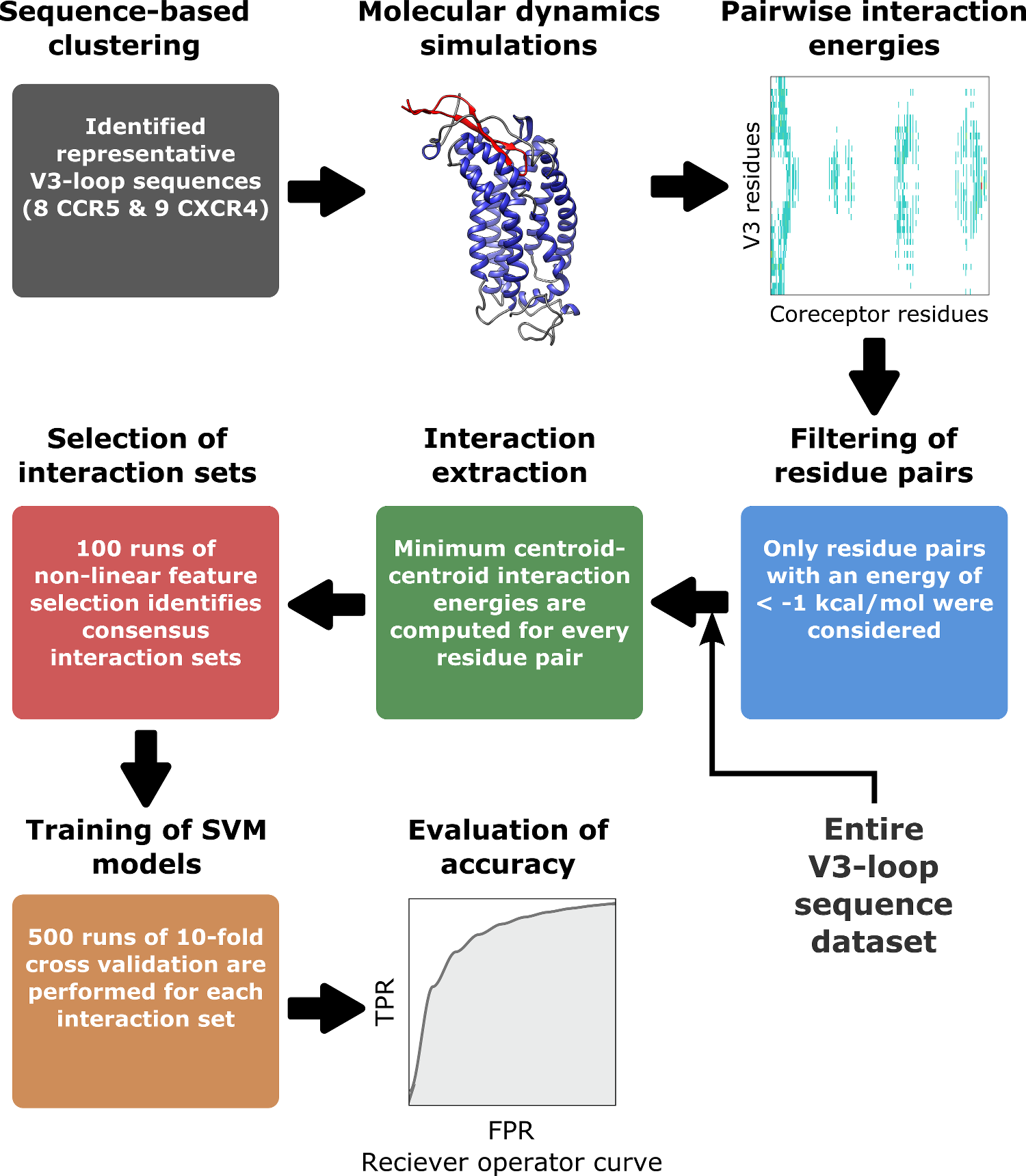
Highly Accurate Structure-Based Prediction of HIV-1 Coreceptor Usage Suggests Intermolecular Interactions Driving Tropism
PLOS ONE
·
09 Feb 2016
·
doi:10.1371/journal.pone.0148974
All
2023
Machine learning models for predicting membranolytic anticancer peptides
Computer Aided Chemical Engineering
·
01 Jan 2023
·
doi:10.1016/b978-0-443-15274-0.50428-5
Data-driven prediction of peptide-MHC binding using oscillations of physicochemical properties
Computer Aided Chemical Engineering
·
01 Jan 2023
·
doi:10.1016/b978-0-443-15274-0.50429-7
2021
Role of Electrostatic Hotspots in the Selectivity of Complement Control Proteins Toward Human and Bovine Complement Inhibition
Frontiers in Molecular Biosciences
·
16 Mar 2021
·
doi:10.3389/fmolb.2021.618068
Data-driven prediction of antiviral peptides based on periodicities of amino acid properties
31st European Symposium on Computer Aided Process Engineering
·
01 Jan 2021
·
doi:10.1016/B978-0-323-88506-5.50312-0
2020
Reassessing enzyme kinetics: Considering protease-as-substrate interactions in proteolytic networks
Proceedings of the National Academy of Sciences
·
24 Jan 2020
·
doi:10.1073/pnas.1912207117
2019
Dynamic Model of Protease State and Inhibitor Trafficking to Predict Protease Activity in Breast Cancer Cells
Cellular and Molecular Bioengineering
·
19 Jun 2019
·
doi:10.1007/s12195-019-00580-5
A nonlinear support vector machine‐based feature selection approach for fault detection and diagnosis: Application to the Tennessee Eastman process
AIChE Journal
·
02 Jan 2019
·
doi:10.1002/aic.16497
2018
Reprint of: Big data approach to batch process monitoring: Simultaneous fault detection and diagnosis using nonlinear support vector machine-based feature selection
Computers & Chemical Engineering
·
01 Aug 2018
·
doi:10.1016/j.compchemeng.2018.10.016
An analysis and evaluation of the WeFold collaborative for protein structure prediction and its pipelines in CASP11 and CASP12
Scientific Reports
·
02 Jul 2018
·
doi:10.1038/s41598-018-26812-8
Big data approach to batch process monitoring: Simultaneous fault detection and diagnosis using nonlinear support vector machine-based feature selection
Computers & Chemical Engineering
·
01 Jul 2018
·
doi:10.1016/j.compchemeng.2018.03.025
Virtual Screening of Chemical Compounds for Discovery of Complement C3 Ligands
ACS Omega
·
15 Jun 2018
·
doi:10.1021/acsomega.8b00606
Optimization of black-box problems using Smolyak grids and polynomial approximations
Journal of Global Optimization
·
03 May 2018
·
doi:10.1007/s10898-018-0643-0
Simultaneous Fault Detection and Identification in Continuous Processes via nonlinear Support Vector Machine based Feature Selection
13th International Symposium on Process Systems Engineering (PSE 2018)
·
01 Jan 2018
·
doi:10.1016/b978-0-444-64241-7.50341-4
2017
AESOP: A Python Library for Investigating Electrostatics in Protein Interactions
Biophysical Journal
·
01 May 2017
·
doi:10.1016/j.bpj.2017.04.005
Princeton_TIGRESS 2.0: High refinement consistency and net gains through support vector machines and molecular dynamics in double-blind predictions during the CASP11 experiment
Proteins: Structure, Function, and Bioinformatics
·
21 Mar 2017
·
doi:10.1002/prot.25274
2016
conSSert: Consensus SVM Model for Accurate Prediction of Ordered Secondary Structure
Journal of Chemical Information and Modeling
·
15 Mar 2016
·
doi:10.1021/acs.jcim.5b00566
Highly Accurate Structure-Based Prediction of HIV-1 Coreceptor Usage Suggests Intermolecular Interactions Driving Tropism
PLOS ONE
·
09 Feb 2016
·
doi:10.1371/journal.pone.0148974
2014
Derivation of ligands for the complement C3a receptor from the C-terminus of C5a
European Journal of Pharmacology
·
01 Dec 2014
·
doi:10.1016/j.ejphar.2014.10.041
Insights into the mechanism of C5aR inhibition by PMX53 via implicit solvent molecular dynamics simulations and docking
BMC Biophysics
·
12 Aug 2014
·
doi:10.1186/2046-1682-7-5
Engineering pre-SUMO4 as efficient substrate of SENP2
Protein Engineering Design and Selection
·
26 Mar 2014
·
doi:10.1093/protein/gzu004
Protein folding and de novo protein design for biotechnological applications
Trends in Biotechnology
·
01 Feb 2014
·
doi:10.1016/j.tibtech.2013.10.008
Forcefield_NCAA: Ab Initio Charge Parameters to Aid in the Discovery and Design of Therapeutic Proteins and Peptides with Unnatural Amino Acids and Their Application to Complement Inhibitors of the Compstatin Family
ACS Synthetic Biology
·
14 Jan 2014
·
doi:10.1021/sb400168u
2013
Forcefield_PTM: Ab Initio Charge and AMBER Forcefield Parameters for Frequently Occurring Post-Translational Modifications
Journal of Chemical Theory and Computation
·
02 Dec 2013
·
doi:10.1021/ct400556v
Princeton_TIGRESS: Protein geometry refinement using simulations and support vector machines
Proteins: Structure, Function, and Bioinformatics
·
22 Nov 2013
·
doi:10.1002/prot.24459
Novel compstatin family peptides inhibit complement activation by drusen-like deposits in human retinal pigmented epithelial cell cultures
Experimental Eye Research
·
01 Nov 2013
·
doi:10.1016/j.exer.2013.07.023
A Predictive Model for HIV Type 1 Coreceptor Selectivity
AIDS Research and Human Retroviruses
·
01 Oct 2013
·
doi:10.1089/aid.2012.0173
2012
The Two Sides of Complement C3d: Evolution of Electrostatics in a Link between Innate and Adaptive Immunity
PLoS Computational Biology
·
27 Dec 2012
·
doi:10.1371/journal.pcbi.1002840
Insights into the Structure, Correlated Motions, and Electrostatic Properties of Two HIV-1 gp120 V3 Loops
PLoS ONE
·
19 Nov 2012
·
doi:10.1371/journal.pone.0049925
De Novo Peptide Design with C3a Receptor Agonist and Antagonist Activities: Theoretical Predictions and Experimental Validation
Journal of Medicinal Chemistry
·
20 Apr 2012
·
doi:10.1021/jm201609k
Clustering of HIV-1 Subtypes Based on gp120 V3 Loop electrostatic properties
BMC Biophysics
·
07 Feb 2012
·
doi:10.1186/2046-1682-5-3
2011
Electrostatic exploration of the C3d–FH4 interaction using a computational alanine scan
Molecular Immunology
·
01 Sep 2011
·
doi:10.1016/j.molimm.2011.05.007
Complement Inhibition by Staphylococcus aureus: Electrostatics of C3d–EfbC and C3d–Ehp Association
Cellular and Molecular Bioengineering
·
18 Aug 2011
·
doi:10.1007/s12195-011-0195-6
The effect of electrostatics on factor H function and related pathologies
Journal of Molecular Graphics and Modelling
·
01 Aug 2011
·
doi:10.1016/j.jmgm.2011.04.010
Electrostatic Similarity Determination Using Multiresolution Analysis
Molecular Informatics
·
01 Aug 2011
·
doi:10.1002/minf.201100002
Automated computational framework for the analysis of electrostatic similarities of proteins
Biotechnology Progress
·
14 Jan 2011
·
doi:10.1002/btpr.541
Is the rigid-body assumption reasonable?
Journal of Non-Crystalline Solids
·
01 Jan 2011
·
doi:10.1016/j.jnoncrysol.2010.05.087
An evaluation of poisson-boltzmann electrostatic free energy calculations through comparison with experimental mutagenesis data
Biopolymers
·
01 Jan 2011
·
doi:10.1002/bip.21644
2010
Electrostatic Clustering and Free Energy Calculations Provide a Foundation for Protein Design and Optimization
Annals of Biomedical Engineering
·
08 Dec 2010
·
doi:10.1007/s10439-010-0226-9
A multifaceted study of stigma/style cysteine-rich adhesin (SCA)-like Arabidopsis lipid transfer proteins (LTPs) suggests diversified roles for these LTPs in plant growth and reproduction
Journal of Experimental Botany
·
28 Jul 2010
·
doi:10.1093/jxb/erq228
Influence of Electrostatics on the Complement Regulatory Functions of Kaposica, the Complement Inhibitor of Kaposi’s Sarcoma-Associated Herpesvirus
The Journal of Immunology
·
20 Jan 2010
·
doi:10.4049/jimmunol.0903261
Solvation effects in calculated electrostatic association free energies for the C3d-CR2 complex, and comparison to experimental data
Biopolymers
·
01 Jan 2010
·
doi:10.1002/bip.21388
2009
A Gain-of-Function Mutation ofArabidopsisLipid Transfer Protein 5 Disturbs Pollen Tube Tip Growth and Fertilization
The Plant Cell
·
01 Dec 2009
·
doi:10.1105/tpc.109.070854